#generate some data
x = np.arange(20)
w = np.array([-3.8,0.11])
y = w[0] + w[1]*x
sigma2 = 0.1
y = y + np.random.normal(0,np.sqrt(sigma2),x.shape[0])
plt.scatter(x,y)
<matplotlib.collections.PathCollection at 0x7fd858496820>
import math
import random
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import plotly.express as px
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LinearRegression
def pretty_print_linear(coefs, intercept, names = None, sort = False):
if names == None:
names = ["X%s" % x for x in range(1,1+len(coefs))]
lst = zip(coefs, names)
if sort:
lst = sorted(lst, key = lambda x:-np.abs(x[0]))
return "%6.3f"%intercept+" + " +" + ".join("%6.3f * %s" % (coef, name)
for coef, name in lst)
This notebook is going to cover a bunch of things related to model evaluation for regression, with a specific focus on linear and polynomial regression:
One important thing we might want to know from a given linear regression model is how any given feature relates to our prediction. We can do this by analyzing the regression coefficients.
#generate some data
x = np.arange(20)
w = np.array([-3.8,0.11])
y = w[0] + w[1]*x
sigma2 = 0.1
y = y + np.random.normal(0,np.sqrt(sigma2),x.shape[0])
plt.scatter(x,y)
<matplotlib.collections.PathCollection at 0x7fd858496820>
def calculateSquaredLoss(X,y,w):
X1 = np.hstack([np.ones([X.shape[0],1]),X])
ypred = np.dot(X1,w)
return 0.5*np.sum(np.power(ypred - y,2))
from sklearn.linear_model import LinearRegression
# fit model - note that LinearRegression's fit function adds the intercept by default
x = np.transpose(np.reshape(x,[1,len(x)]))
y = np.transpose(np.reshape(y,[1,len(y)]))
lr = LinearRegression()
lr.fit(x,y)
print(pretty_print_linear(lr.coef_,lr.intercept_))
-4.005 + 0.112 * X1
The key thing to remember is that a linear regressioncoefficient tells us how much our modeled outcome changes with a one-unit change in $x$.
If we have multiple predictors, i.e. ${\bf x}$, then we append to this above statement holding all other predictors fixed.
Regression models do not care what the $x$ and $y$ are. Just because $x$ predicts $y$, then, isn't a causal statement, because it is equally valid to flip the model around and say that $y$ causes $x$. There are at least two ways to get to causation: 1) to use social theory, and 2) to use causal inference. Both come with many assumptions, we will discuss this later in the semetser.
As we discuss elsewhere in class, our model learns parameters based on training data that is simply a sample from a population. Thus, one reasonable question to ask is, how do I know if the relationship between my $x$ and my $y$, as measured by the regression coefficients/parameters, is "real"? There are many ways to answer this question. Perhaps the most obvious and well-established is to place a confidence interval on your parameter estimate using statistical inference. As always StatQuest has a nice explanation of this. We will discuss this in more detail at some point later in class. But another approach to thinking about this for ML, where we often care mainly about prediction, is that this relationship is "real" if it is useful in making predictions. We're going to stick with that one for now.
Your interpretation of your features depends on the scale of both $x$ and $y$. Sometimes, to make interpretation easier, you may want to rescale your input, or your outputs. Some points to remember:
Assuming you have an intercept in your model, and no interaction terms, rescaling changes your interpretation of a linear model, but not the actual fit. Here is a useful explanation of why.
data_size = 500
data = pd.DataFrame({"covid_deaths_per_person_in_state": [random.gauss(.01,1) for _ in range(data_size)],
"covid_cases_per_state": [random.gauss(1000,10) for _ in range(data_size)]
})
data['local_news_covid_articles'] = data.covid_deaths_per_person_in_state + data.covid_cases_per_state
mod = LinearRegression(fit_intercept=False).fit(data[['covid_deaths_per_person_in_state',
'covid_cases_per_state']],
data.local_news_covid_articles)
mod.coef_
array([1., 1.])
Exercise: In this example, what is more important, deaths or cases? Do our regression coefficients tell us that?
Changing the scale of your $y$ changes your interpretation as well! We discussed an example in class, and there is one on your programming assignment. There's some useful resources on the internet for this point if you want to learn more, in particular, here is one useful exploration for how to interpret linear coefficients with a logged outcome.
Finally, we will see models where rescaling/standardizing your variables is critical for model performance. Linear regression is not one of them, but other regression and classification approaches definitely are impacted by the scale of your input variables. More on this later!
The most obvious place this comes up is in the context of using categorical variables in your regression.
Consider a regression model where we are trying to predict the amount of water you drink in a day. We have one feature, whether or not you have exercised:
data = pd.DataFrame({"exercised" : random.choices(['yes','no'], k=500)})
## Mini-quiz ... what is this code doing?
data['water'] = 10 + 3*(data.exercised == "yes").astype(int)
OK! Let's try to fit that!
mod = LinearRegression(fit_intercept=False).fit(data[['exercised']],
data.water)
--------------------------------------------------------------------------- ValueError Traceback (most recent call last) /var/folders/rf/4jz25sh1733_8b28phhhrlfh0000gn/T/ipykernel_51448/833097770.py in <module> ----> 1 mod = LinearRegression(fit_intercept=False).fit(data[['exercised']], 2 data.water) ~/opt/anaconda3/lib/python3.8/site-packages/sklearn/linear_model/_base.py in fit(self, X, y, sample_weight) 660 accept_sparse = False if self.positive else ["csr", "csc", "coo"] 661 --> 662 X, y = self._validate_data( 663 X, y, accept_sparse=accept_sparse, y_numeric=True, multi_output=True 664 ) ~/opt/anaconda3/lib/python3.8/site-packages/sklearn/base.py in _validate_data(self, X, y, reset, validate_separately, **check_params) 579 y = check_array(y, **check_y_params) 580 else: --> 581 X, y = check_X_y(X, y, **check_params) 582 out = X, y 583 ~/opt/anaconda3/lib/python3.8/site-packages/sklearn/utils/validation.py in check_X_y(X, y, accept_sparse, accept_large_sparse, dtype, order, copy, force_all_finite, ensure_2d, allow_nd, multi_output, ensure_min_samples, ensure_min_features, y_numeric, estimator) 962 raise ValueError("y cannot be None") 963 --> 964 X = check_array( 965 X, 966 accept_sparse=accept_sparse, ~/opt/anaconda3/lib/python3.8/site-packages/sklearn/utils/validation.py in check_array(array, accept_sparse, accept_large_sparse, dtype, order, copy, force_all_finite, ensure_2d, allow_nd, ensure_min_samples, ensure_min_features, estimator) 744 array = array.astype(dtype, casting="unsafe", copy=False) 745 else: --> 746 array = np.asarray(array, order=order, dtype=dtype) 747 except ComplexWarning as complex_warning: 748 raise ValueError( ~/opt/anaconda3/lib/python3.8/site-packages/pandas/core/generic.py in __array__(self, dtype) 1991 1992 def __array__(self, dtype: NpDtype | None = None) -> np.ndarray: -> 1993 return np.asarray(self._values, dtype=dtype) 1994 1995 def __array_wrap__( ValueError: could not convert string to float: 'no'
Exercise: What happened?
... OK, let's fix the issue, naively
data['exercised_yes'] = (data.exercised == "yes").astype(int)
data['exercised_no'] = (data.exercised == "no").astype(int)
mod = LinearRegression(fit_intercept=False).fit(data[['exercised_yes','exercised_no']],
data.water)
mod.coef_
array([13., 10.])
Hmph... that worked, but ... that doesn't match our true function! Note, however, that 13-10 = 3, which is what our expected result would have been for a single coefficient.
Exercise: what happened? (don't peek!)
... note that our regression function as specified is:
$ y = w_0 + w_1*exercised + w_2 no\_exercise + \epsilon$
Now, note that: $exercised = 1 - no\_exercise$, so we can rewrite that as:
$ y = w_0 + w_1*exercised + w_2 - w_2 exercised + \epsilon $, and so:
$ y = (w_0 + w_2) + (w_1-w_2) exercised + \epsilon$
Exercise: How many $w_1$ and $w_2$s can we find that solve this equation?
Note: the closed form solution wouldn't even have worked here! (Because we can't invert this matrix). But sklearn still gives us an output! Here is an explanation of why, you're not responsible for understanding that explanation, though.
OK, so, now we know that we should arbitrarily pick either yes or no and drop that column. Here, it makes sense to drop no, although in many cases this choice is arbitrary. Having done so, our model becomes:
$ y = w_0 + w_1*exercised + \epsilon$
Exercise: What does $w_0$ now represent?
Finally, note that all of this logic extends to categorical variables, which we "one-hot encode" too!
Aside: not all transformations are linear rescaling. Exercise: What is an example of a transformation that might help us make predictions?
To evaluate our model's predictions, we have to be able to actually make those predictions. At a high level, we make predictions by simply subbing in the $x$ value in the test data into our trained model. That is, assume we have trained a model $f_{\hat{w}}$ (using the notation in the UW course). Then to make a prediction on a test point, we evaluate $f_{\hat{w}}$ at $x$, i.e. we compute $f_{\hat{w}}(x)$.
It can in many cases be useful to evaluate a single model, but typically it is very hard to know how well you are making predictions if you look at one and only one model combined with one and only one set of features. Exercise: why?
In class, we generalized regression, following the end of Chapter 1 from Hunter Schafer's book. You are responsible for understanding this generalized equation.
At a high level, though, there are a few different things we can do to try to make different/better predictions:
Note that, as we discussed in class, 1 and 2 are tightly intertwined, in that some models (e.g. polynomial regression) implicitly "add" to our features
We're going to do this "actual model comparison" in two ways. In the first, we're going to assume we know the true function. If we know the true function, we can assess the bias/variance tradeoff of our model by direct evaluation of model bias and variance!
In this section, we are going to:
sklearnOne way to think about ML is, as we have discussed, in terms of trying to estimate (conditional probability) functions. A good way to think about this is to think about there being some true function f that we want to learn.
Sadly, we don't know the functional form (linear? polynomial?) or the parameters of the model (in terms of what we have seen so far, what are the ws?).
Our goal in ML is to learn f given only one random training set that gives us some x and some y, where y is, as noted further below, the output of f(x) plus some irreducible noise.
Exercises
In lecture, we discussed how the source of error in our estimation of f(x) using training data comes from one of three places:
y_i = f(x_i) + epsilon_i, and it is hopeless to learn the epsilon_i since its random and does not depend on the inputRemember this picture for bias/variance:
from IPython.display import Image
from IPython.core.display import HTML
Image(url= "https://miro.medium.com/max/384/1*dxzjw6RMg6myHwk1HEKdKQ.png",
width=400)
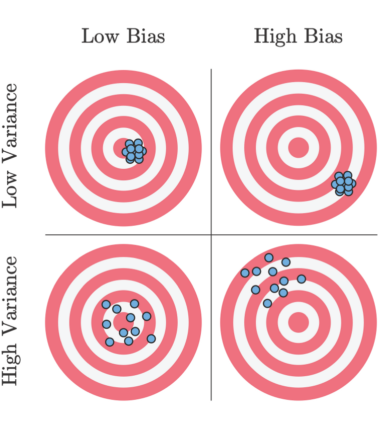
There is a fundamental tradeoff between bias and variance that depends on how complex your model is. Very simple models (with what we've seen so far, models with few parameters) have high bias since your true function is usually not constant, but have low variance since they generally don't have the complexity to fit the noise of the specific dataset you got. Very complex models (high degree polynomials) have low bias since you can get a descent approximation of the true function in expectation, but have high variance since they have the capabilities of fitting the noise in the data.
This section has some code examples to demonstrate how this bias variance tradeoff occurs with different model complexities using synthetic data.`
MIN_X = -1
MAX_X = 6
def f(x):
"""
True function f(x) (unknown to us)
"""
return 2 * x - 1.4 * x ** 2 + .2 * x ** 3
px.line(pd.DataFrame({"x": np.linspace(MIN_X,MAX_X, 30),
"y" : [f(x) for x in np.linspace(MIN_X,MAX_X, 30)]}),
x="x",y="y",
height=400,width=400)
The generate_data function below will generate x values uniformly at random in [min_x, max_x] and then assign the values using the function f and Gaussian noise with mean 0 and variance noise_sd.
def generate_data(n,min_x=0,max_x=1, noise_sd = .1):
"""
Creates and returns a SFrame with n randomly drawn examples
"""
xs = [random.uniform(min_x, max_x) for _ in range(n)] # generate n numbers uniform at random from [0, 1]
ys = [f(x) + random.gauss(0, noise_sd) for x in xs] # evaluate f at each x and add Gaussian noise (mean=0, variance=.1)
return pd.DataFrame({'x': xs, 'y': ys})
def polynomial_features(data, col, deg):
"""
Given a dataset, creates a polynomial expansion of the input with
the given name to the given degree.
Returns the dataset and the list of column names
"""
data_copy = data.copy()
if deg == 0:
data_copy[col + '0'] = 0
columns = [col + '0']
else:
columns = []
for i in range(1, deg + 1): # +1 to include deg
col_name = col + str(i)
data_copy[col_name] = data_copy[col] ** i
columns.append(col_name)
return data_copy, columns
Below is an example dataset we might observe.
data = generate_data(n=50)
data, columns = polynomial_features(data, 'x', deg=3)
print(columns)
data.head()
['x1', 'x2', 'x3']
| x | y | x1 | x2 | x3 | |
|---|---|---|---|---|---|
| 0 | 0.242160 | 0.409650 | 0.242160 | 0.058641 | 0.014201 |
| 1 | 0.341757 | 0.602137 | 0.341757 | 0.116798 | 0.039916 |
| 2 | 0.472779 | 0.671901 | 0.472779 | 0.223520 | 0.105676 |
| 3 | 0.829565 | 0.760769 | 0.829565 | 0.688178 | 0.570889 |
| 4 | 0.417905 | 0.494490 | 0.417905 | 0.174645 | 0.072985 |
If you run it multiple times, you would most likely get different values. Exercise: Why?
This feels kind of unrealistic - we only have one training dataset! True. But let's say our training dataset is a set of tweets sent today, and we train a model to predict number of retweets. There's a lot of randomness in that sample, and if we sampled again tomorrow, it would likely lead to a (slightly) different model!
The following function will learn some given number of models using random samples from our training set. Exercise: how are we getting those random samples?
This is technique of approximating getting new datasets from the underlying distribution with only our one dataset is known as resampling.
We will use each random (re)sample of our training set to train a model
def learn_models(data, features, num_models):
"""
Trains num_models models on random subsets of the given data.
Returns the list of learned models.
"""
models = []
for i in range(num_models):
# get a random sample of the datat
random_sample_from_X, _, random_sample_from_y, _= train_test_split(data[features].values,data.y.values,test_size=0.4)
model = LinearRegression().fit(random_sample_from_X,random_sample_from_y)
models.append(model)
return models
models = learn_models(data, columns, 3)
for mod in models:
print(pretty_print_linear(mod.coef_,mod.intercept_))
-0.078 + 2.713 * X1 + -2.887 * X2 + 1.146 * X3 0.099 + 1.294 * X1 + -0.106 * X2 + -0.512 * X3 0.001 + 1.884 * X1 + -0.772 * X2 + -0.317 * X3
These models are all different, even though they're estimating the same f!!!
Exercises
learn_models to make them all (almost) the same?f if f(x) = 2 * x - 1.4 * x ** 2 + .2 * x ** 3 + x**4?Now, we want to think about how different forms we could specify for our guess at f lead to different levels of bias and variance. To do that, we're going to train a bunch of models, and use them all to make predictions at a bunch of values of x. Let's call each model $f_{\hat{w}}(x)$, representing the fact that they are all approximations of f based on different parameters $\hat{w}$. We're then going to plot the range of the predictions across all models to get a sense of variance, and $\mathbb{E_{\hat{w}}}[f_{\hat{w}}(x)]$ to get a sense of the bias.
def find_predictions_range(models, model_degree, num_eval_points, min_x=0, max_x=1):
"""
Compares the predictions of the given models at num_eval_points between min_x and max_x.
This gives a description of how spread the predictions are at a particular point x.
If the predictions are all over the place, then the range of predictions will be high.
Returns:
* A list of points evaluated (of length num_eval_points)
* A list of tuples containing information about the predictions at each eval point.
Each tuple contains the minimum prediction, mean prediction, and max prediction for that input
"""
new_xs = np.linspace(min_x, max_x, num_eval_points) # just like range, but allows for fractional steps
new_data = pd.DataFrame({'x': new_xs})
new_data, columns = polynomial_features(new_data, 'x', model_degree)
all_predictions = [model.predict(new_data[columns].values) for model in models]
intervals = []
for i in range(len(new_xs)):
# get predictions from each model for this input i
preds_i = [preds[i] for preds in all_predictions]
# find the mean, min, and max
intervals.append((min(preds_i), np.mean(preds_i), max(preds_i)))
return new_xs, intervals
x, model_pred_intervals = find_predictions_range(models, 3, 10, MIN_X,MAX_X)
print(x)
print("_________")
print(model_pred_intervals[0])
[-1. -0.22222222 0.55555556 1.33333333 2.11111111 2.88888889 3.66666667 4.44444444 5.22222222 6. ] _________ (-6.823340291037015, -3.317104640622015, -0.7897900273220814)
def plot(data, new_xs, intervals,min_x=0,max_x=1):
"""
Plots the data and range of predictions to demonstrate the bias and variance of the model
"""
# magic to separate list of [(low, mean, high)] to list of lows, list of means, list of highs
lows, means, highs = zip(*intervals)
# plot the range bands for predictions
plt.fill_between(new_xs, lows, highs, facecolor='#E8F8F8', edgecolor='#c3e2e2',
label='Range of predictions')
# plot the average predictions (won't perfectly match f because it's an approximation of E_train[hat{f}]
plt.plot(new_xs, means, color="r", label='Average hat{f}')
# plot the true f
plt.plot(new_xs, [f(x) for x in new_xs], color='g', label='True f')
# plot the datapoints
plt.scatter(data['x'], data['y'], c='black', s=10, label='Data')
# modify axis to make it look nice and add label
axes = plt.gca()
axes.set_xlim([min_x, max_x])
axes.set_ylim([data['y'].min() - 0.5, data['y'].max() + 0.5])
plt.legend(loc='upper left', bbox_to_anchor=(1, 1))
plt.show()
This is the main function to run our experiments. See the cell aftewards how to call it.
def bias_variance(num_data, num_models, degree_expansions, num_eval_points=200, min_x=0, max_x=1,noise_sd=.1):
"""
Demonstrates the bias-variance tradeoff for each degree in degree_expansions using
one randomly generated dataset of num_dat samples with value some true f(x) + noise.
For each degree, trains num_models models using features up to that degree. It then uses
those models to make predictions for the num_eval_points between 0 and 1. It records
these predictions and visually plots the range of predictions for each x.
"""
print('Generating data')
data = generate_data(num_data,min_x,max_x,noise_sd)
for degree_expansion in degree_expansions:
print('Evaluating model with degree ' + str(degree_expansion))
# generate data with
data, columns = polynomial_features(data, 'x', degree_expansion)
models = learn_models(data, columns, num_models)
new_xs, intervals = find_predictions_range(models, degree_expansion, num_eval_points,min_x,max_x)
plot(data, new_xs, intervals,min_x,max_x)
bias_variance(num_data= 30,
num_models = 100,
degree_expansions = [0, 1, 2, 5, 10],
num_eval_points = 50,
min_x=MIN_X,
max_x=MAX_X,
noise_sd=1)
Generating data Evaluating model with degree 0
Evaluating model with degree 1
Evaluating model with degree 2
Evaluating model with degree 5
Evaluating model with degree 10
The way to read each of the graphs is:
fExercises
noise_sd to be really large?f - would the same models still be the best?num_data -i.e. increase the size of the dataset?We've already seen some of this stuff in Lecture 4 with our introduction to why we need a train/test split. But there's more to understand here!
Like last time, we're going to run through some lecture notes to build out the intuition first, and then play in the notebook and notes next week (Tuesday)